I have prepared this post as documentation for a speech I will give on November 12th with my colleagues of Grupo-R madRid. In our previous meeting Jesús Herranz gave us a good introduction on survival models, but he reserved the best stuff for his workshop on random forests for survival, which happened in our recent VII R-hispano users group congress -maybe the best event about R in Spain.
I had recently prepared an introduction on survival models focused on business settings -as a contribution to the applicability of these wonderful models outside the more frequent biomedical field- as my first techie blog post -in spanish (!). In the meantime I also found these absolutely gReat tutorials on survival for business by Dayne Batten, and now the idea is to quickly apply part of the lessons learnt at Jesus’ workshop to the churn dataset.
We will actually follow only half of Jesus’ modeling proposals, in particular the libraries randomForestSRC -gReatest thanks to Hemant Ishwaran, and ggRandomForests for visualization.
Let’s go into the code. It should work on any R environment, but please be very careful with some hints for installing the above libraries. If you find any problem please write a comment!
You can download all this content and its code from an Rmarkdown file in my github.
Setup your R environment
Be sure to change your working directory:
setwd("d:/survival")
Load (and install if necessary) the required libraries (but see below special requirement about the randomForestSRC library).
list.of.packages <- c("survival",
"caret",
"glmnet",
"rms",
"doParallel",
"risksetROC")
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) install.packages(new.packages)
library(survival, quietly = TRUE)
library(caret, quietly = TRUE)
library(glmnet, quietly = TRUE)
library(rms, quietly = TRUE)
library(risksetROC, quietly = TRUE)
library(doParallel, quietly = TRUE)
registerDoParallel(detectCores() - 1 ) ## registerDoMC( detectCores()-1 ) in Linux
detectCores()
options(rf.cores = detectCores() - 1,
mc.cores = detectCores() - 1) ## Cores for parallel processing
Read more about doParallel.
By default, doParallel uses multicore functionality on Unix-like systems and snow functionality on Windows. Note that the multicore functionality only runs tasks on a single computer, not a cluster of computers.
randomForestSRC package allows parallelization but the library binaries are different for Windows and Linux, so you must go to Hemant Ishwaran’s rfsrc page and download the zip file and install it as source.
My system is a windows 7 machine, so I am using one version of this zip. Use the appropriate one for your platform. Please verify it loads in your system.
install.packages("http://www.ccs.miami.edu/~hishwaran/rfsrc/randomForestSRC_1.6.0.zip",
repos = NULL,
type = "source")
library(randomForestSRC)
And only after this you can install ggRandomForests, with very useful plotting functions for the random forests objects created with randomForestSRC.
install.packages("ggRandomForests",
repos = 'http://cran.us.r-project.org') #since you had source before
library(ggRandomForests)
Load and explore the data about churn
I found these Churn data (artificial based on claims similar to real world) suggested at stackoverflow question. Data are part of UCI machine learning training sets, also more quickly found at http://www.sgi.com/tech/mlc/db/.
nm <- read.csv("http://www.sgi.com/tech/mlc/db/churn.names",
skip = 4,
colClasses = c("character", "NULL"),
header = FALSE,
sep = ":")[[1]]
dat <- read.csv("http://www.sgi.com/tech/mlc/db/churn.data",
header = FALSE,
col.names = c(nm, "Churn"),
colClasses = c("factor",
"numeric",
"factor",
"character",
rep("factor", 2),
rep("numeric", 14),
"factor"))
# test data
test <- read.csv("http://www.sgi.com/tech/mlc/db/churn.test",
header = FALSE,
col.names = c(nm, "Churn"),
colClasses = c("factor",
"numeric",
"factor",
"character",
rep("factor", 2),
rep("numeric", 14),
"factor"))
This is a quick exploration of training dataset. You have 3333 unique customer id’s (phone numbers), account.length is the age (time dimension), which seems to be months, but I am not totally sure, and you have 15% drop-outs, which is a quite high churn rate but then consider we have >10 years span.
dim(dat)
summary(dat)
length(unique(dat$phone.number))
hist(dat$account.length)
table(dat$Churn)/nrow(dat)*100
## [1] 3333 21
## state account.length area.code phone.number
## WV : 106 Min. : 1 408: 838 Length:3333
## MN : 84 1st Qu.: 74 415:1655 Class :character
## NY : 83 Median :101 510: 840 Mode :character
## AL : 80 Mean :101
## OH : 78 3rd Qu.:127
## OR : 78 Max. :243
## (Other):2824
## international.plan voice.mail.plan number.vmail.messages
## no :3010 no :2411 Min. : 0.0
## yes: 323 yes: 922 1st Qu.: 0.0
## Median : 0.0
## Mean : 8.1
## 3rd Qu.:20.0
## Max. :51.0
##
## total.day.minutes total.day.calls total.day.charge total.eve.minutes
## Min. : 0 Min. : 0 Min. : 0.0 Min. : 0
## 1st Qu.:144 1st Qu.: 87 1st Qu.:24.4 1st Qu.:167
## Median :179 Median :101 Median :30.5 Median :201
## Mean :180 Mean :100 Mean :30.6 Mean :201
## 3rd Qu.:216 3rd Qu.:114 3rd Qu.:36.8 3rd Qu.:235
## Max. :351 Max. :165 Max. :59.6 Max. :364
##
## total.eve.calls total.eve.charge total.night.minutes total.night.calls
## Min. : 0 Min. : 0.0 Min. : 23.2 Min. : 33
## 1st Qu.: 87 1st Qu.:14.2 1st Qu.:167.0 1st Qu.: 87
## Median :100 Median :17.1 Median :201.2 Median :100
## Mean :100 Mean :17.1 Mean :200.9 Mean :100
## 3rd Qu.:114 3rd Qu.:20.0 3rd Qu.:235.3 3rd Qu.:113
## Max. :170 Max. :30.9 Max. :395.0 Max. :175
##
## total.night.charge total.intl.minutes total.intl.calls total.intl.charge
## Min. : 1.04 Min. : 0.0 Min. : 0.00 Min. :0.00
## 1st Qu.: 7.52 1st Qu.: 8.5 1st Qu.: 3.00 1st Qu.:2.30
## Median : 9.05 Median :10.3 Median : 4.00 Median :2.78
## Mean : 9.04 Mean :10.2 Mean : 4.48 Mean :2.77
## 3rd Qu.:10.59 3rd Qu.:12.1 3rd Qu.: 6.00 3rd Qu.:3.27
## Max. :17.77 Max. :20.0 Max. :20.00 Max. :5.40
##
## number.customer.service.calls Churn
## Min. :0.00 False.:2850
## 1st Qu.:1.00 True. : 483
## Median :1.00
## Mean :1.56
## 3rd Qu.:2.00
## Max. :9.00
##
## [1] 3333

And about the test set. You have exactly 1667 rows, exactly half of the training set.
summary(test);dim(test)
Random Forests for Survival
Random Forests (RF) is a machine learning technique which builds a large number of decision trees that:
- are based on bootstrap samples. Each tree is based on a random sample with replacement of all observations.
- each tree division is based on a random sample of predictors.
- There is no prunning, trees are as long as possible, they are not “cut”.
For building each RF tree a part of the observations is not used (37% aprox.). This is called out-of-bag -OOB- sample and is used for a honest estimate of the model predictive capability.
Random Survival Forest (RSF) is a class of survival prediction models, those that use data on the life history of subjects (the response) and their characteristics (the predictor variables). In this case, it extends the RF algorithm for a target which is not a class, or a number, but a survival curve. The library is actually so clever that given a particular target it automatically selects the relevant algorith. There are four families of random forests:
regression forests for continuous responses
classification forests for factor responses
Survival forests for right-censored survival settings
competing risk survival forests for competing risk scenarios
RF is now a standard to effectively analyze a large number of variables, of many different types, with no previous variable selection process. It is not parametric, and in particular for survival target it does not assume the proportional risks assumption.
rfsrc requires all data to be either numeric or factors. So you must filter out character, date or other types of variables. Survival object requires a numeric (0/1) target, and in my R environment I have had problems to input factors into the analysis, so I quickly drop those variables (in previous analyses they were not any relevant). And we quickly -and dirtly- convert into dummies (0/1) two relevant factors and the target (Churn).
dat$phone.number <- NULL
dat$state <- NULL
dat$area.code <- NULL
dat$international.plan <- as.numeric(dat$international.plan) - 1
dat$voice.mail.plan <- as.numeric(dat$voice.mail.plan) - 1
dat$Churn <- as.numeric(dat$Churn) - 1
summary(dat)
You use rfsrc() to build a RF model with the following parameters:
formula : response variable and predictors
data : data frame containing data
ntree: total number of trees
mtry: number of variables entering in each division as candidates. By default sqrt(p) in classification and survival and p/3 in regression.
nodesize : minimum number of observations in a terminal node (survival, 3).
nsplit : number of random points to explore in continous predictors.
importance = T : prints out variable importance ranking (if not use importance=“noneâ€).
proximity = T : to compute this metric.
Handling factors is not that easy. See the “Allowable data types and issues related to factors” part in rfsrc documentation.
Let’s try a simple RF model with 50 trees and nsplit 2.
out.rsf.1 <- rfsrc(Surv(account.length, Churn) ~ . ,
data = dat,
ntree = 50,
nsplit = 2)
out.rsf.1
## Sample size: 3333
## Number of deaths: 483
## Number of trees: 50
## Minimum terminal node size: 3
## Average no. of terminal nodes: 352.3
## No. of variables tried at each split: 4
## Total no. of variables: 16
## Analysis: RSF
## Family: surv
## Splitting rule: logrank *random*
## Number of random split points: 2
## Error rate: 13.75%
Computation is intensive (though we have requeste a very simple model and we do not have too many data). rsfrc() permutesall values of all variables in all trees. However, at least for me, the parallelization implementation works flawlessly and gives output very quickly.
The $importance object contains variables importance, in same order as in input dataset. We sort it out to show a ranking and use ggRandomForestses to plot this object. This library uses ggplot2, so you can easily retouch these plots.
imp.rsf.1 <- sort(out.rsf.1$importance,
decreasing = T)
imp.rsf.1
plot(gg_vimp(out.rsf.1))
## international.plan total.day.charge
## 0.1192592 0.0820343
## total.day.minutes number.customer.service.calls
## 0.0696993 0.0639611
## voice.mail.plan total.eve.minutes
## 0.0288920 0.0222369
## total.eve.charge number.vmail.messages
## 0.0214636 0.0106841
## total.intl.minutes total.intl.calls
## 0.0106587 0.0093293
## total.intl.charge total.night.charge
## 0.0086888 0.0046463
## total.night.minutes total.eve.calls
## 0.0026219 0.0009676
## total.day.calls total.night.calls
## 0.0006847 -0.0006018
This is the variable importance plot

Predictive ability of RSF
To compute a numeric prediction per case, we sum the risk output estimate along all times. This is equivalent to a risk score, so that higher values correspond to observations with higher observed risk, lower survival. These predictions can be based on all trees or only on the OOB sample.
In RSF, error rate is defined as = 1 – C-index. C-index means that the higher the survival, the lower the risk. Let’s check C-index.
length(out.rsf.1$predicted.oob)
## [1] 3333
head(out.rsf.1$predicted.oob)
## [1] 24.412 6.364 9.577 58.879 52.422 31.528
sum.chf.oob = apply(out.rsf.1$chf.oob , 1, sum)
head(sum.chf.oob)
## [1] 24.412 6.364 9.577 58.879 52.422 31.528
rcorr.cens(out.rsf.1$predicted.oob,
Surv(dat$account.length, dat$Churn))["C Index"]
## C Index
## 0.1371
err.rate.rsf = out.rsf.1$err.rate[ out.rsf.1$ntree ]
err.rate.rsf
## [1] 0.1375
rcorr.cens(-out.rsf.1$predicted.oob,
Surv(dat$account.length, dat$Churn))["C Index"]
## C Index
## 0.8629
Towards an optimal RFS
An essential parameter is the number of trees. To compute the best number of trees you use importance=“none†so that you do not use unnecessary computing, and set a sufficiently high number of trees. This depends as you could see by default values, on the number of predictor variables. In our example, with just a few variables, a value of a few hundreds looks enough. We then use gg_error() of ggRandomForests to plot the results across the number of trees. You choose the point (minimum number) where plot converges into a minimum. If it does not converge, try with a higher number of trees.
out.rsf.3 <- rfsrc( Surv(account.length, Churn) ~ . ,
data = dat,
ntree = 200,
importance = "none",
nsplit = 1)
out.rsf.3
plot(gg_error(out.rsf.3))
## Sample size: 3333
## Number of deaths: 483
## Number of trees: 200
## Minimum terminal node size: 3
## Average no. of terminal nodes: 365.4
## No. of variables tried at each split: 4
## Total no. of variables: 16
## Analysis: RSF
## Family: surv
## Splitting rule: logrank *random*
## Number of random split points: 1
## Error rate: 13.97%

Predictive ability applied to test data: C Index
Let’s do as usual: apply our model to the test data and check for predictive ability.
First of all, remember to make same mods as we did to the training to test set!!!
test$phone.number <- NULL
test$state <- NULL
test$area.code <- NULL
test$international.plan <- as.numeric(test$international.plan) - 1
test$voice.mail.plan <- as.numeric(test$voice.mail.plan) - 1
test$Churn <- as.numeric(test$Churn) - 1
summary(test)
We apply the computed model to the test set using, as usual, predict. We then check the C_Index as before.
pred.test.fin = predict( out.rsf.3,
newdata = test,
importance = "none" )
rcorr.cens(-pred.test.fin$predicted ,
Surv(test$account.length, test$Churn))["C Index"]
## C Index
## 0.8596
Predictive ability. ROC generalizations
risksetROC library provides functions to compute the equivalent to a ROC curve and its associated Area Under Curve (AUC) in a time-dependent context.
This is precisely the greatest advantage: you can compute predictive ability at specific time points or intervals. Let’s see predictive ability of OOB samples (in training set) at median time. We must be careful about method as, depending on assumptions, this can be different (see documentation), but let’s assume we meet Cox’s proportional hazards assumption.
w.ROC = risksetROC(Stime = dat$account.length,
status = dat$Churn,
marker = out.rsf.3$predicted.oob,
predict.time = median(dat$account.length),
method = "Cox",
main = paste("OOB Survival ROC Curve at t=",
median(dat$account.length)),
lwd = 3,
col = "red" )
w.ROC$AUC

## [1] 0.8095
For risksetROC to compute AUC along an interval you use risksetAUC using tmax (maximum time). You get a very nice plot of AUC across time. This is still OOB samples.
w.ROC = risksetAUC(Stime = dat$account.length,
status = dat$Churn,
marker = out.rsf.3$predicted.oob,
tmax = 250)

Let’s do the same for test data.
w.ROC = risksetAUC(Stime = test$account.length,
status = test$Churn,
marker = pred.test.fin$predicted,
tmax = 220,
method = "Cox")
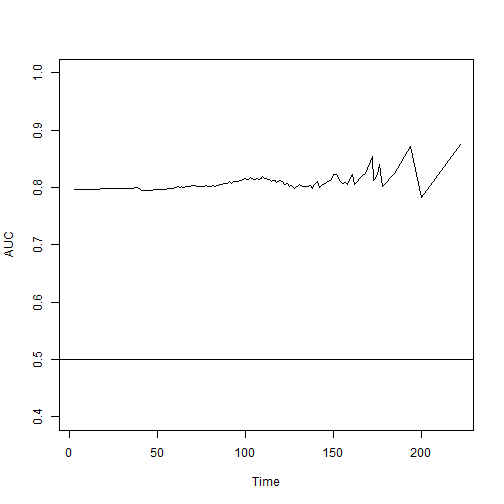
And with a plot, at good local maximum prediction time, 190.
w.ROC = risksetROC(Stime = test$account.length,
status = test$Churn,
marker = pred.test.fin$predicted,
predict.time = 190,
method = "Cox",
main = paste("OOB Survival ROC Curve at t=190"),
lwd = 3,
col = "red" )
w.ROC$AUC

w.ROC$AUC
## [1] 0.8269
And with a plot, at maybe best prediction time, 220 (?????).
w.ROC = risksetROC(Stime = test$account.length,
status = test$Churn,
marker = pred.test.fin$predicted,
predict.time = 220,
method = "Cox",
main = paste("OOB Survival ROC Curve at t=220"),
lwd = 3,
col = "red" )

w.ROC$AUC
## [1] 0.8138
Conclusions
- randomForestSRC is a fabulous library for all tasks related to Random Forests computing, and it has a wondeRful implementation for survival targets.
- I have found some issues for handling factors that I shall explore, so that you can input factors directly into the algorithm (without converting into dummies).
- RSF gives important advantages over traditional (classification) RF when you have survival (time dependent) targets. In particular you can check predictive capability and its changes across time, and many other useful results for obtaining knowledge of your analysis. All this maintaing the predictive ability of the traditional classification models.
I hope this encourages you to try these models and these libraries for your analyses. Feel free to comment your results, or problems if you found them!


Thank you for this informative article. I am a bit confused on how we predict “time to churn” for active customers (churn=FALSE) if such data is already used in training.
The train dataset you have used contains both customers who churned after ‘n’ days and customers who are still active (churn=FALSE). So how can we accurately predict the “time to churn” (survival) for such customers since that data is actually used for creating the model?
Ideally I would use only churned customer data for train/test and then predict survival for un-churned customers. How would one do that with rfsrc?
LikeLike
Hi, I am not sure if I understand your question. Both train and test datasets contain churners and active users. This particular split (which I have not done, since I take datasets directly from the sgi.com repository) looks like the usual 70/30 random split of the data at a single point in time. This split is useful for validating the model, though I recognize that in practical settings what you have is a train dataset at a moment in time and a test set *later in time*.
Is this what you intend? I understand that your problem is that you want to make predictions of survival for customers *after* you have computed the model. Then these data might not be suitable for that purpose.
And for your question “So how can we accurately predict the “time to churn” (survival) for such customers since that data is actually used for creating the model?” this is the same issue as when you want to predict from a random forest, regression model or whatever.
Let me check again and I will redo the script and will provide an example on prediction.
LikeLike
Your response makes things clear to me. The “point in time” snapshot is what I was missing. It makes sense that the train dataset is perhaps an earlier point in time data and test data is later in time. Thank you.
LikeLike
Sorry, but I had one other question.
The survival probabilities for test data are consistently very high. I would have assumed that the survival probability at least for those observations that are uncensored would be pretty low given their event has already occurred. But that doesn’t seem the case. Am I missing something in how these probabilities are interpreted?
LikeLike
At what time are you predicting? This must be clearly defined, survival probability depends on time. Can you give an example of the code you have used? Or an example of point you find particularly high.
By the way I am very very happy that this small work is found useful by someone out there. Thanks for commenting and asking!!
LikeLike
Here’s the code I am using from the default pbc dataset of randomForestSRC package.
##load data
data(pbc, package = “randomForestSRC”)
pbc.trial % dplyr::filter(!is.na(treatment))
pbc.test % dplyr::filter(is.na(treatment))
##build model
rfsrc_pbc <- rfsrc(Surv(days, status) ~ .,
data = pbc.trial,
na.action = "na.impute")
##test model – test data contains un-censored data
test.pred.rfsrc <- predict(rfsrc_pbc,
pbc.test,
na.action="na.impute")
##check times
test.pred.rfsrc$time.interest
##compare survival probabilities
ndf2 <- as.data.frame(test.pred.rfsrc$survival[,11]) #survival probability for 191 days
y2 <- cbind(ndf2, status=pbc.test$status)
mean(dplyr::filter(y2, status==1)[,1]) #expect this to be low because event has occurred- but is 0.95
mean(dplyr::filter(y2, status==0)[,1]) #expect this to be high in comparison – is 0.98
LikeLiked by 1 person
Hi, jjreddick
I see you use pbc data from the randomForestSRC library. Please check what is said about this dataset in the library documentation:
Click to access randomForestSRC.pdf
A total o f424 PBC patients during 10 year interval randomized placebo controlled trial. So treatment = 1 / 2 is placebo or not. The first 312 cases in the data set participated in the randomized trial and contain largely complete data.
I think you had the idea that those with treatment == NA are those in a “test” set. But what happens is that they simply have most of data missing. Check
head(pbc[is.na(pbc$treatment),], n = 50)
Please consider a more traditional train/test split, only with the 312 complete data:
pbc2 <- pbc[!is.na(pbc$treatment), ]
smp_size <- floor(0.70 * nrow(pbc2))
## set the seed to make your partition reproductible
set.seed(123)
train_ind <- sample(seq_len(nrow(pbc2)), size = smp_size)
pbc.train <- pbc2[train_ind, ]
pbc.test <- pbc2[-train_ind, ]
nrow(pbc.train)
nrow(pbc.test)
##build model
rfsrc_pbc <- rfsrc(Surv(days, status) ~ .,
data = pbc.train)
##test model – test data contains un-censored data
test.pred.rfsrc <- predict(rfsrc_pbc,
pbc.test)
##check times
test.pred.rfsrc$time.interest
hist(test.pred.rfsrc$time.interest)
Are these predictions more reasonable for you now?
LikeLike
Thank you again Pedro – and I hate to take up any more of your time. One last follow up if you don’t mind since I may not have clarified my question earlier.
Following from your code:
##test model – test data contains un-censored data
test.pred.rfsrc <- predict(rfsrc_pbc,
pbc.test,
na.action="na.impute") #added this so I get results for all test rows
test.pred.rfsrc$time.interest #check times for which survival probabilities exist e.g. 2nd column is survival probabilities for 77th day
head(test.pred.rfsrc$survival[,2]) #check survival probabilities at 77th day
res <- data.frame(surv_prob=test.pred.rfsrc$survival[,2], status=pbc.test$status) #compare survival prob with actual status
head(res, 12)
surv_prob status
1 0.9826833 1 #since status=1 I would have expected survival probability returned by the model to be very low since event has already occurred.
2 0.9956333 1
3 1.0000000 1 #again status=1 but model is telling us 100% chance that this row will survive for 77 days
4 0.9997500 1
5 0.9954833 1
6 0.9241571 1
7 0.9991333 1
8 0.9896833 1
9 0.9989333 1
10 1.0000000 0
11 0.9962000 0
12 0.9854333 1
In this result set the survival probability is very high at 77th day for those rows whose status=1 indicating event has already occurred. The probabilities for other days are also relatively high. Am I mis-reading these probability values.
LikeLike
I might have been wrong in the way I was thinking about the values in test.pred.rfsrc$time.interest – it seems this is an observation’s total number of days. I was thinking it returns total number of days an observation will survive from this point onwards. Is that right?
LikeLike
Sorry jjreddick, I missed your two last comments. Yes, time.interest is predicted survival time at the time of using the computed survival model. It is in the same units as the time variable that is used for computing the model (could be days, minutes, months, whatever unit in the input dataset).
I have seen that rows at the beginning are status = 1, and those at the end are 0. Then this can be easy to check (but please note I have no idea of this particular dataset)
If you use
head(pbc.test)
head(test.pred.rfsrc$time.interest)
tail(pbc.test)
tail(test.pred.rfsrc$time.interest)
You see how predicted survival time is low for those who have status = 1. Yes, the event might have happened, but this is days. For instance see the riskiest case, case 3 in test, who is 25594 days old (70 years). A survival time of 77 days is extremely low, when days in treatment are 1012 (7%).
What you have in
head(test.pred.rfsrc$survival)
Are estimated probabilities of survival at specifi moments in time. Note how different are these times from moment 10, for instance. This is very rich information, but of course it depends on the particular event and input variables.
Happy again you found this exercise useful.
Pedro
LikeLike
Hi,
I recently used this package for the first time and it seems great. I too encountered the problem you discussed with factors. The problem is that in my case the factors had a number of different levels, so they couldn’t be ‘binarized’. One option is to use one-hot-encoding but that would drastically increase the feature space since my covariates are all factors.
I worked around the problem by simply coding the factors as numbers, running the algorithm, and then coding the variables back.
Not a reasonable approach, I know. Did you happen to explore this problem with factors further?
Thanks for the great article. The validation/visualization part was especially useful.
Rohail
LikeLike